Timothy Cody

Data Scientist with a Master's in Analytics and 3 years in experince delevering analytical reporting and data driven insights.
Skills: Python, R, SQL, Power BI, Tableau
View My LinkedIn Profile
Deep Learning for DDI Prediction
Project description: Multiclass classification project for predicting drug-drug interactions from chemical structures using deep learning.
Details: The importance of predicting drug-drug interactions (DDIs) cannot be overstated, given the risks they pose to patient safety and efficacy of medication management. DDIs are a common cause of adverse drug reactions (ADRs), particularly in elderly patients requiring polypharmacy. In fact, the prevalence of DDIs in polypharmacy patients ranges from 20-40%, highlighting the urgent need to develop effective strategies to mitigate these risks. Computational modeling has emerged as a promising approach to predicting potential DDIs, enabling clinicians and drug developers to identify potential interactions before they occur. Accurately predicting DDIs can lead to better clinical outcomes, reduced healthcare costs, and improved patient quality of life. Therefore, continued research and development in this area is critical to advancing the field of drug development and improving patient care.
This project aimed to develop a computational model to predict potential drug-drug interactions (DDIs) based solely on molecular structure. Accurately predicting DDIs can help mitigate the risks associated with polypharmacy and improve patient outcomes. Therefore, this study sought to build upon existing research and identify the most effective molecular representation for predicting DDIs.
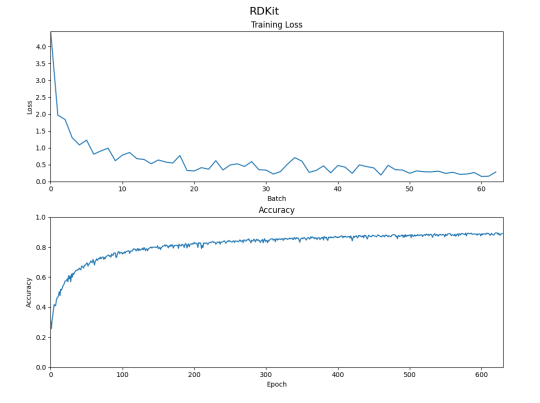
To achieve this goal, several fingerprint representations were evaluated, including the ECFP4 fingerprint, Morgan fingerprint, and RDKit fingerprint. The performance of these fingerprints was compared based on their ability to accurately predict DDIs in a large database of drug pairs. The RDKit fingerprint was found to be the most effective at retaining the most information and achieving the best performance on the test set.
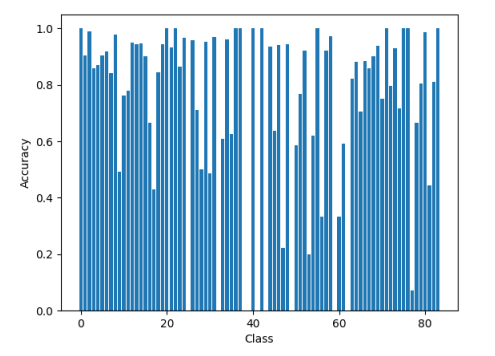
The final model developed in this study achieved a training accuracy of over 90%, which is on par with existing methods available in the literature. However, further improvements are possible by making changes to the drug representation or the structure of the network. Despite achieving high overall accuracy, the model struggled with some classes of DDIs, highlighting the need for further research to improve accuracy in these areas.
In summary, this project contributes to the development of effective strategies for mitigating the risks associated with DDIs. By accurately predicting potential interactions, clinicians and drug developers can improve patient outcomes, reduce healthcare costs, and enhance the quality of life for patients. Moreover, this study highlights the importance of selecting the appropriate molecular representation for predicting DDIs and provides insights for future research in this area.